Clustering of HIV-1 sequences among men who have sex with men in Beijing, China
Simon Frost, M.A. D.Phil.
Dept. of Veterinary Medicine, and Institute of Public Health
University of Cambridge
Co-authors and funders
- Vanderbilt
- Marcia Kalish
- Han-Zhu Qian
- Lu Yin
- Joseph Conrad
- Sten Vermund
- China CDC
- Yi Feng
- Yuhua Ruan
- Yiming Shao
- Hui Xing
- Beijing CDC
- Yuejuan Zhao
- HJF/DAIDS/NIH
- Hans Spiegel
- Funding
- NIH/NIAID
- UK MRC
Introduction
- While HIV is endemic in many regions, in some populations, HIV is just emerging
- The HIV epidemic is emerging among men who have sex with men in Beijing, China
- What might we learn about this emerging epidemic from combining phylogenetic and behavioural data?
Individual-level data and clustering
- How can we combine individual-level data with sequence data?
- Highlighted as a challenge by Frost et al., Epidemics (2015)
- Most studies have reduced sequence data to whether an individual is clustered or not
- Two individuals cluster if their viral sequences are within a threshold distance
- May also be other criteria, e.g. bootstrap/posterior probability support
Why cluster?
- Computational reasons: easier to fit clusters of related sequences than all of them
- e.g. UK HIV Drug Resistance Database c. 100,000 sequences
- (but can lead to biased estimates of phylodynamic parameters; see Bethany Dearlove's talk)
- Populations are structured; clusters may reveal subepidemics with higher transmission rates
- Can use standard statistical approaches
Why not cluster?
- Loss of information
- Problems with clustering thresholds
- Evolutionary rate. The rate may change with transmission rate, and vary by subtype.
- Time since infection. What happens with a mixture of individuals at different stages of disease?
- Sampling. Sampling rate can change over time and space
- Intermediate number of infections may vary by stage of infection
- Choice of threshold. Too small - not enough clusters. Too large - everyone is clustered.
Methods for Prevention Packages Program study
- NIH funded program for reducing HIV transmission
- Vanderbilt/China CDC study in Beijing, China
- Recruited from March 2013 to March 2014
- Subset of 356 individuals with HIV pol data, who self identify as MSM, completed a behavioural questionnaire
Modeling the transmission of HIV in Beijing

Lou et al. PLoS ONE (2014)
Projections of HIV in Beijing

Lou et al. PLoS ONE (2014)
Subtyping process
- We used a variety of subtyping tools:
- Rega (V2, V3)
- SCUEAL
- COMET
- jpHMM
- STAR
- RIP
- BLAST
- Exploratory phylogenetic analysis
Subtypes
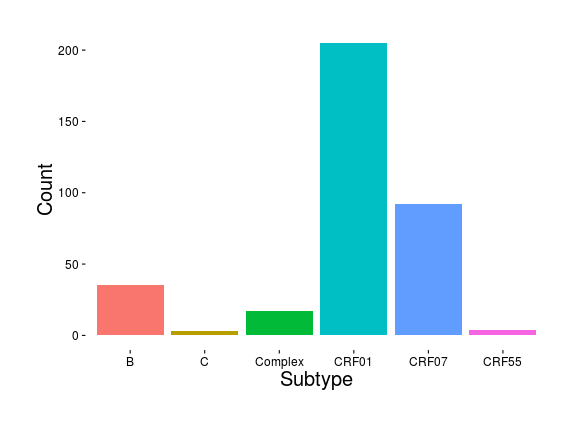
Frost et al., draft.
Subtypes and internet use
Clustering and minimum distance
- We calculated pairwise distances between all sequences in the sample
- Sergei Kosakovsky Pond's TN93 program
- For each sequence, selected the minimum of these distances
- Large differences between subtypes
Differences between subtypes
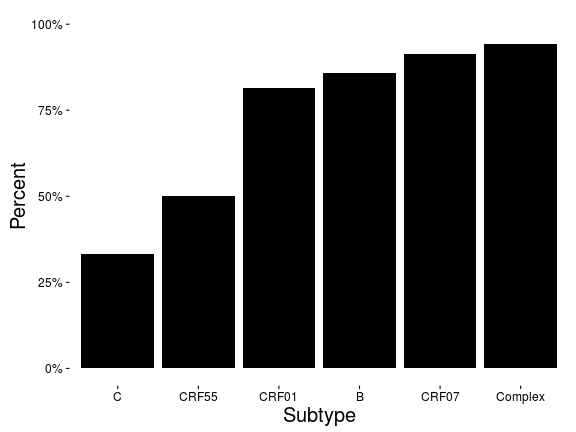
Patterns of clustering (<1%)

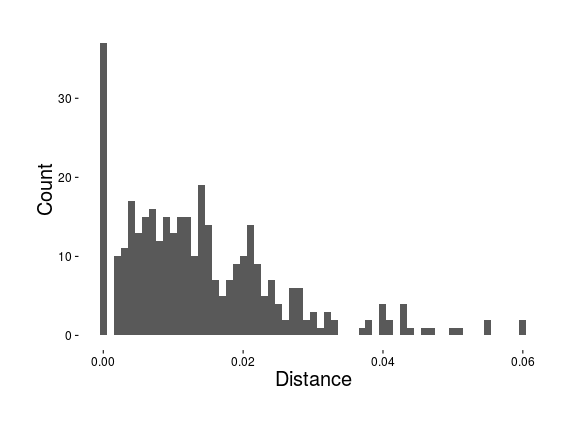
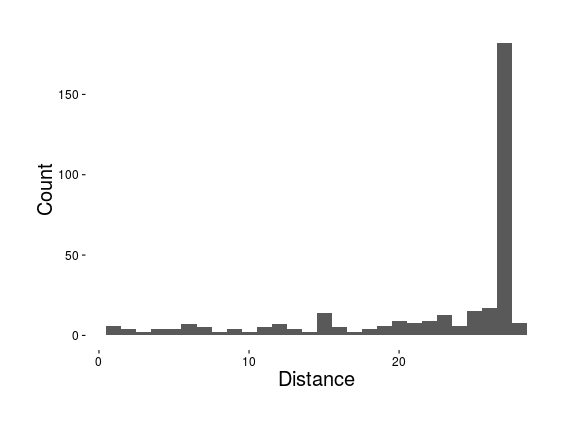
Distribution of distances
Clustering with 'background' sequences
- Downloaded partial pol sequences from Genbank, at least 1000 bp long
- 108,135 'background' sequences
- 424 were sampled from MSM in Beijing
- 108,135 'background' sequences
- Calculated the (TN93) distance from the sample sequences to the background sequences
'Background' clustering

Modeling clustering
- Problem:
- On the one hand, we have a clear excess of very close (<0.1%) clustering
- On the other, the distances vary greatly by subtype
- Sampling times are too close together to reliably estimate evolutionary rate
- Solution:
- Take minimum distance as an outcome
- Model this as a zero adjusted Gamma (ZAGA) model
Zero adjusted Gamma
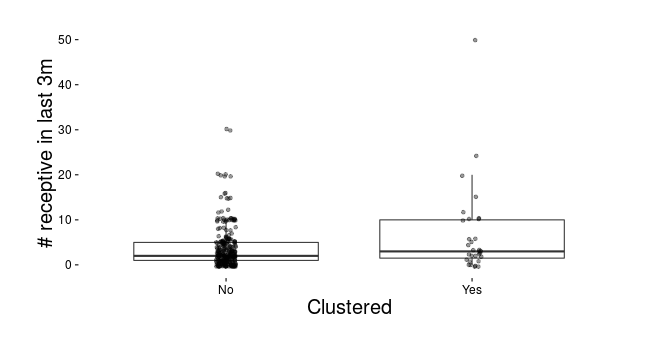
Clustering and receptive intercourse
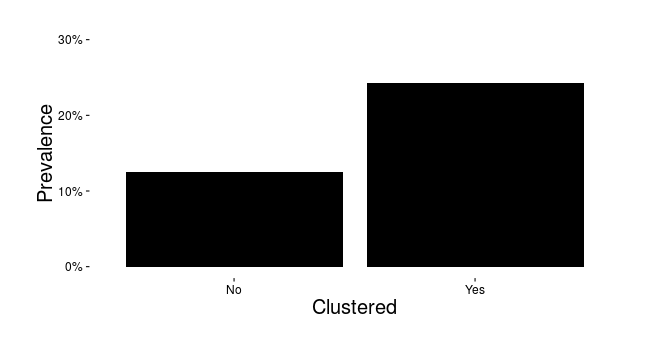
Clustering and active syphilis
Distance and years resident in Beijing

Who are people clustering with?
- In addition to calculating the minimum distance, we can examine the characteristics of the individual who is linked to each
- We calculated the number of ambiguous nucleotides ('mixtures') that did not affect the amino acid sequence for each individual
- Crude measure of within-host diversity, which increases during infection
Links per alter

Lessons from modeling
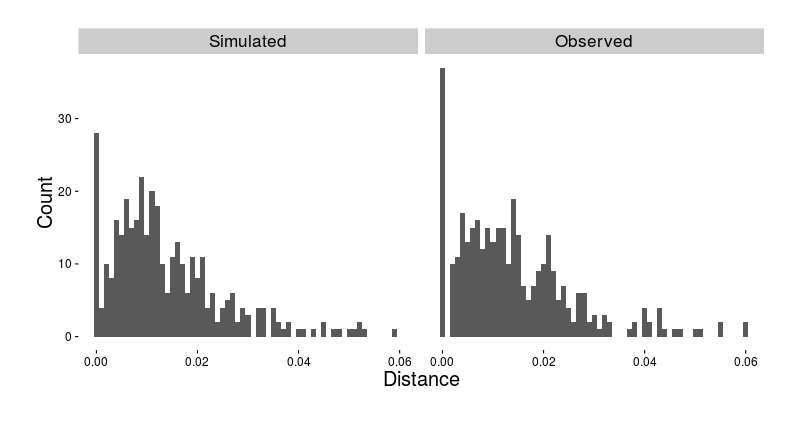
- We combined the mathematical model of Lou et al. (2014) with the phylodynamics framework of Volz (2012)
- Simulated trees with 356 taxa, with the same sampling times as the observed data
- Calculated the minimum cophenetic distance for each taxon
Minimum distance in simulation
Next steps
- Retrospective analysis of HIV sequence data collected by China CDC
- Better behavioral data collection?
- No obvious patterns between subtypes
- Model development
- Immigration of infecteds
- More structure in the population
Conclusions
- Exponential epidemic of HIV-1 among MSM in Beijing
- Particularly CRF07_BC, perhaps linked to finding partners on the internet
- Mixture of potentially direct transmission and looser association with risk groups
- Not seen in 'well mixed' models of transmission
- Demographic data reinforce the importance of movement of people
- Such 'late' emergence is not unique
- Manila, Philippines
Thanks!
 sdwfrost@gmail.com
sdwfrost@gmail.com
 @sdwfrost
@sdwfrost
 http://github.com/sdwfrost
http://github.com/sdwfrost
 sdwfrost@gmail.com
sdwfrost@gmail.com
 http://github.com/sdwfrost
http://github.com/sdwfrost

